Mechanochemical Models
Actomyosin dynamics
During cell migration, the actin cytoskeleton takes on distinct structures in different regions of the cell. We use particle-based computer simulations to study the molecular mechanisms that give rise to these emergent properties of the cytoskeleton. Our models account for biophysical interactions between filamentous actin and non-muscle myosin II. Our simulations reveal the spontaneous formation of actin asters from spatially homogenous initial conditions. We are investigating how the model responds in a gradient to spatial chemical gradients. Interestingly, spatial regulation of motor stiffness leads to a time-dependent behavior of the actomyosin network, in which actin asters continue to spontaneously form and dissociate (see Movie).
Computational Modeling of Cerebral Cavernous Malformations
Defects in the proper formation of blood vessels and capillaries lead to many diseases. In particular, cerebral cavernous malformations (CCMs) are clusters of leaky, dilated capillaries causing seizures, stroke, and neurological deficits. CCMs occur in 0.5-1.5% of the population, and are only treatable through complex and often risky surgical intervention. A deficiency in any of three CCM proteins (CCM1, CCM2, or CCM3) is sufficient to cause the disease. The Johnson lab has demonstrated using genetic approaches that CCM1, 2 and 3 are required for initiating proper tube formation. The CCM proteins are known to regulate the mechanical properties of the cell by controlling the activity of RhoA, a small GTPase that activates actomyosin based contractility and regulates the actin cytoskeleton. We use mathematical modeling to gain a mechanistic understanding of how the CCM proteins regulate the intracellular signaling system that in turn controls the mechanical properties of the cell. Importantly, the computational tools developed for these investigations will not only help to predict therapeutic targets for CCM treatment but also should significantly impact other fields where multicellular formation is a crucial aspect of the physiological process.
Molecular motors
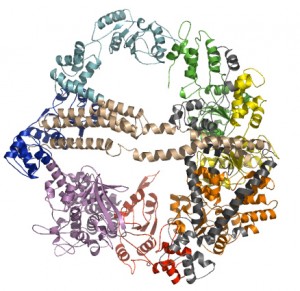
Our work on molecular motors focuses on understanding the mechanisms used by these molecules to convert free energy, stored in the forms of chemical bonds or ion gradients, into mechanical work. Most recenlty, our work has focused on dynein. Homology modeling is being used to generate an accurate structure of this molecule that will then be used in coarse-grained molecular dynamics simulations to understand the conformational changes that are used to generate forces.
We are also interested in developing numerical methods for studying force generation by molecular motors. Recently we have extended our methods to include the biophysical properties of the linkage that connects the motor to its cargo.
Dynein Stepping
Coupling of Structural and Kinetic Models of Molecular Motor Dynein
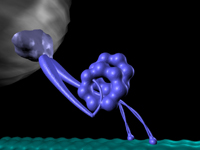
Using a coarse-grained structural model for two-headed dynein and a hybrid of molecular dynamics and Monte Carlo simulations, we explore the mechanical properties of dynein stepping, including: a structurally biased search for binding sites, step-size distribution, mechanical modulation of transition rates, and the dependence of these features on the orientation and flexibility of the motor’s interaction with the microtubule. By simulating the processive motion of dynein, we demonstrate the consistency of our structural model and earlier kinetic studies with experimental observations.
Serohijos AWR, Tsygankov D, Liu S, Elston TC, Dokholyan NV. Multiscale approaches for studying energy transduction in dynein. Phys Chem Chem Phys. 2009 Jun 28; 11(24): 4840-50.
(Pubmed | Journal)
Tsygankov D, Serohijos AW, Dokholyan NV, Elston TC. Kinetic models for the coordinated stepping of cytoplasmic dynein. J Chem Phys. 2009 Jan 14;130(2):025101
(Pubmed | Journal)
Energy transduction in dynein
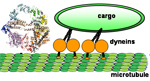 Intracellular active transport is driven by ATP-hydrolyzing motor proteins that move along cytoskeletal filaments. In particular, the microtubule-associated dynein motor is involved in the transport of organelles and vesicles, the maintenance of the Golgi, and mitosis. However, unlike kinesin and myosin, the mechanism by which dynein converts chemical energy into mechanical force remains largely a mystery, due primarily to the lack of a high-resolution molecular structure. Using homology modeling and normal mode analysis, we propose a complete atomic structure and a mechanism for force generation by the motor protein dynein.
Intracellular active transport is driven by ATP-hydrolyzing motor proteins that move along cytoskeletal filaments. In particular, the microtubule-associated dynein motor is involved in the transport of organelles and vesicles, the maintenance of the Golgi, and mitosis. However, unlike kinesin and myosin, the mechanism by which dynein converts chemical energy into mechanical force remains largely a mystery, due primarily to the lack of a high-resolution molecular structure. Using homology modeling and normal mode analysis, we propose a complete atomic structure and a mechanism for force generation by the motor protein dynein.
Serohijos AW, Chen Y, Ding F, Elston TC, Dokholyan NV. A structural model reveals energy transduction in dynein. Proc Natl Acad Sci U S A. 2006 Dec 5; 103(49):18540-5
(Pubmed | Journal)
The role of motor-cargo linkage in molecular motors
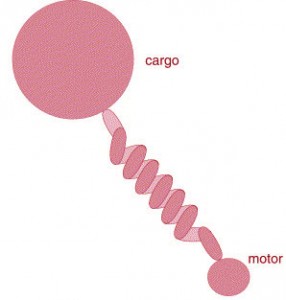 The development of single-molecule techniques for measuring the biophysical properties of molecular motors has motivated the use of mathematical models to elucidate the mechanisms used by these proteins for force generation. Processive motors, such as kinesin, myosin, and dynein, are used for transporting vesicles within cells and for force generation during processes such as mitosis. In general, processive molecular motors consist of two head domains and move along microtubule or actin polymers. The heads interact with specific binding sites along the polymer track and are connected through a stalk that extends from the neck region of each head. The stalk ends in a tail domain that is used to connect the motor to its cargo or to adjacent polymers. Until recently most of the models of molecular motor function did not take into account the biophysical properties of the stalk region that connects the motor to its cargo. However, it is becoming increasingly clear that this linkage can have important consequences on the performance of the motor.
The development of single-molecule techniques for measuring the biophysical properties of molecular motors has motivated the use of mathematical models to elucidate the mechanisms used by these proteins for force generation. Processive motors, such as kinesin, myosin, and dynein, are used for transporting vesicles within cells and for force generation during processes such as mitosis. In general, processive molecular motors consist of two head domains and move along microtubule or actin polymers. The heads interact with specific binding sites along the polymer track and are connected through a stalk that extends from the neck region of each head. The stalk ends in a tail domain that is used to connect the motor to its cargo or to adjacent polymers. Until recently most of the models of molecular motor function did not take into account the biophysical properties of the stalk region that connects the motor to its cargo. However, it is becoming increasingly clear that this linkage can have important consequences on the performance of the motor.
Fricks J, Wang H, Elston TC. A numerical algorithm for investigating the role of the motor-cargo linkage in molecular motor-driven transport. J Theor Biol. 2006 Mar 7; 239(1):33-48
(Pubmed | Journal)
Further Publications
Goedecke DM, Elston TC. A model for the oscillatory motion of single dynein molecules. J Theor Biol. 2005 Jan 7; 232(1):27-39
(Pubmed | Journal)
