Signal Transduction
 A main focus of our laboratory is to use computational and mathematical methods to discover and understand control mechanisms used to regulate signaling pathways. In general, signaling pathways are highly nonlinear and inherently noisy systems. They often contain multiple feedback and feedforward loops and share common functional components. Therefore, the broad questions we seek to address are: What biological functions do feedback and feedforward loops provide? Is noise reduction important for maintaining signaling integrity? How is pathway specificity achieved?
A main focus of our laboratory is to use computational and mathematical methods to discover and understand control mechanisms used to regulate signaling pathways. In general, signaling pathways are highly nonlinear and inherently noisy systems. They often contain multiple feedback and feedforward loops and share common functional components. Therefore, the broad questions we seek to address are: What biological functions do feedback and feedforward loops provide? Is noise reduction important for maintaining signaling integrity? How is pathway specificity achieved?
 To answer these questions, we have chosen to study the mating response pathway of yeast S. cerevisiae. This system is arguably the best characterized signaling pathway of any eukaryote, and it has long served as a prototype for hormone, neurotransmitter, and sensory response systems in humans. We have developed an interdisciplinary research program that combines computational modeling with experimental analysis. Both deterministic and stochastic models of G-protein and protein kinase activity are being developed and validated against experimental data from the laboratories of Drs Henrik Dohlman and Beverly Errede. The models are used to generate testable hypotheses that define the next generation of experiments.
To answer these questions, we have chosen to study the mating response pathway of yeast S. cerevisiae. This system is arguably the best characterized signaling pathway of any eukaryote, and it has long served as a prototype for hormone, neurotransmitter, and sensory response systems in humans. We have developed an interdisciplinary research program that combines computational modeling with experimental analysis. Both deterministic and stochastic models of G-protein and protein kinase activity are being developed and validated against experimental data from the laboratories of Drs Henrik Dohlman and Beverly Errede. The models are used to generate testable hypotheses that define the next generation of experiments.
Chemotropic Growth in Pheromone Gradient
Right click to download matlab files:
Gradient chamber simulation
Colony simulation
Finding mate simulation
Competition for mate simulation
Quantification of Biosensor Data
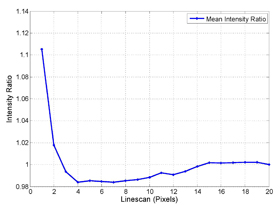
The Hahn lab develops biosensors for studying adhesion and motility in eukaryotic cells. In vivo imaging of biosensors leads to a plethora of data. Analyzing this data for novel biological observations is often challenging due to cell to cell heterogeneity, intra-cell heterogeneity and low signal-to-noise ratio. Our current research involves development of quantification techniques that ameliorate these concerns. This is particularly challenging at the cell edge, where the signal to noise ratio can be at its worst. Software has been developed to automatically analyze biosensor data at the cell edge. This converts the 2D image into a profile of activity relative to the edge. Currently we are working on how to correct for artifacts at the cell edge in ratio images.
The procedure creates a matrix of activities at the edge. We are studying how to correlate these with velocities and cluster the linescans to derive quantitative information re protein activity during motility.
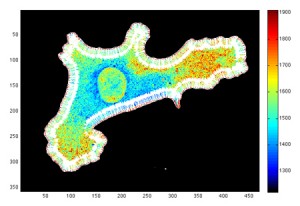
Figure 3
Graphical representation of the Line Scan Matrix. Each element of the matrix has a ratio (or intensity value) and is indexed by its depth along the line scan and its position along the edge (line scan number).
Further Publications
Zuo P, Picher M, Okada SF, Lazarowski ER, Button B, Boucher RC, Elston TC. Mathematical model of nucleotide regulation on airway epithelia: Implications for airway homeostasis. J Biol Chem. 2008 Sept 26; 283(39):26805-26819
(Pubmed | Journal)
Hao N, Nayak S, Behar M, Shanks RH, Nagiec MJ, Errede B, Hasty J, Elston TC, Dohlman HG. Regulation of cell signaling dynamics by the protein kinase-scaffold Ste5. Mol Cell. 2008 Jun 6; 30(5):649-56
(Pubmed | Journal)
Violin JD, DiPilato LM, Yildirim N, Elston TC, Zhang J, Lefkowitz RJ. β2-adrenergic receptor signaling and desensitization elucidated by quantitative modeling of real time cAMP dynamics. J Biol Chem. 2008 Feb 1; 283(5):2949-61
(Pubmed | Journal)
Behar M, Dohlman HG, Elston TC. Kinetic insulation as an effective mechanism for achieving pathway specificity in intracellular signaling networks. Proc Natl Acad Sci U S A. 2007 Oct 9; 104(41):16146-51. Epub 2007 Oct 3.
(Pubmed | Journal)
Behar M, Hao N, Dohlman HG, Elston TC. Mathematical and computational analysis of adaptation via feedback inhibition in signal transduction pathways. Biophys J. 2007 Aug 1; 93(3):806-21
(Pubmed | Journal)
Hao N, Behar M, Elston TC, Dohlman HG. Systems biology analysis of G protein and MAP kinase signaling in yeast. Oncogene. 2007 May 14; 26(22):3254-66
(Pubmed | Journal)
Hao N, Behar M, Parnell SC, Torres MP, Borchers CH, Elston TC, Dohlman HG. A systems-biology analysis of feedback inhibition in the Sho1 osmotic-stress-response pathway. Curr Biol. 2007 Apr 17; 17(8):659-67
(Pubmed | Journal)
Wang X, Hao N, Dohlman HG, Elston TC. Bistability, stochasticity, and oscillations in the mitogen-activated protein kinase cascade. Biophys J. 2006 Mar 15; 90(6):1961-78
(Pubmed | Journal)
Yildirim N, Hao N, Dohlman HG, Elston TC. Mathematical modeling of RGS and G-protein regulation in yeast. Methods Enzymol. 2004; 389:383-98
(Pubmed | Journal)
Hao N, Yildirim N, Wang Y, Elston TC, Dohlman HG. Regulators of G protein signaling and transient activation of signaling: experimental and computational analysis reveals negative and positive feedback controls on G protein activity. J Biol Chem. 2003 Nov 21; 278(47):46506-15
(Pubmed | Journal)